 |
gstlal-inspiral
0.4.2
|
The purpose of this page is to investigate the effect of changing the autocorrelation length.
Three identical NSBH - nonspinning analyses were completed. The only change between each run was to the autocorrelation lentgh.
# Template bank parameters
LOW_FREQUENCY_CUTOFF = 30.0
HIGH_FREQUENCY_CUTOFF = 2048.0
NUM_SPLIT_TEMPLATES = 200
OVERLAP = 20
BANK_PROGRAM = pycbc_geom_nonspinbank
APPROXIMANT = TaylorF2
# controls triggers
IFOS = H1 L1 V1
START = 966384015
STOP = 967384015
TAG = tito-bank-1401
WEBDIR = ~/public_html/MDC/NSBH/Summer2014/recolored/nonspin/$(START)-$(STOP)-$(TAG)
NUMBANKS = 4
PEAK = 0
AC_LENGTH = 1401
SAMPLES_MIN = 512
SAMPLES_MAX_256 = 512
# additional options, e.g.,
#ADDITIONAL_DAG_OPTIONS = "--blind-injections BNS-MDC1-WIDE.xml"
# Injections
# The seed is the string before the suffix _injections.xml
# Change as appropriate, whitespace is important
INJECTIONS := HL-INJECTIONS_976731_NSBHMDCINJ_SET1_T4-966384015-5184000.xml HL-INJECTIONS_976765_NSBHMDCINJ_SET1_T2-966384015-5184000.xml
FAR_INJECTIONS := HL-FAR_INJECTIONS_976731_NSBHMDCINJ_SET1_T4-966384015-5184000.xml HL-FAR_INJECTIONS_976765_NSBHMDCINJ_SET1_T2-966384015-5184000.xml
comma:=,
INJECTION_REGEX = $(subst $(space),$(comma),$(INJECTIONS))
FAR_INJECTION_REGEX = $(subst $(space),$(comma),$(FAR_INJECTIONS))
# Segment and frame type info
SEG_SERVER=https://segdb.ligo.caltech.edu
LIGO_FRAME_TYPE='T1200307_V4_EARLY_RECOLORED_V2'
VIRGO_FRAME_TYPE='T1300121_V1_EARLY_RECOLORED_V2'
LIGO_SEGMENTS="$*:CBC-MDC1_SCIENCE_EARLY_RECOLORED:2"
VIRGO_VETOES="V1:INJECTION_INSPIRAL,V1:INJECTION_BURST"
LIGO_VETOES="$*:DMT-INJECTION_INSPIRAL,$*:DMT-INJECTION_BURST"
H1_CHANNEL=LDAS-STRAIN
L1_CHANNEL=LDAS-STRAIN
V1_CHANNEL=h_16384Hz
CHANNEL_NAMES:=--channel-name=H1=$(H1_CHANNEL) --channel-name=L1=$(L1_CHANNEL) --channel-name=V1=$(V1_CHANNEL)
# Get some basic definitions
include Makefile.offline_analysis_rules
#
# Workflow
#
all : dag
#NSBH_216_nospin_blue_30early.xml.gz:
bank_no_spin.xml.gz :
#gsiscp sugar-dev1.phy.syr.edu:/home/spxiwh/aLIGO/comp_costing/bank_sizes/new_runs/NSBH_216_nospin_blue/30_earlyaligo/NSBH_216_nospin_blue_30early.xml.gz .
gsiscp h2.atlas.aei.uni-hannover.de:/home/tito/projects/spin_search/nsbh_mdc1/bank_no_spin/bank_no_spin.xml.gz .
$(INJECTIONS):
gsiscp ldas-pcdev1.ligo-wa.caltech.edu:/home/spxiwh/aLIGO/MDC/MDC1/inj_sets/inj_set_1/"{$(INJECTION_REGEX)}" .
$(FAR_INJECTIONS):
gsiscp ldas-pcdev1.ligo-wa.caltech.edu:/home/spxiwh/aLIGO/MDC/MDC1/inj_sets/inj_set_1/"{$(FAR_INJECTION_REGEX)}" .
%_split_bank.cache : bank_no_spin.xml.gz
mkdir -p $*_split_bank
gstlal_bank_splitter --output-path $*_split_bank --approximant $(APPROXIMANT) --bank-program $(BANK_PROGRAM) --output-cache $@ --overlap $(OVERLAP) --instrument $* --n $(NUM_SPLIT_TEMPLATES) --sort-by mchirp --add-f-final --max-f-final $(HIGH_FREQUENCY_CUTOFF) --group-by-chi $<
plots :
mkdir plots
$(WEBDIR) :
mkdir -p $(WEBDIR)
tisi.xml :
ligolw_tisi --instrument=H1=0:0:0 --instrument=H2=0:0:0 --instrument=L1=0:0:0 --instrument=V1=0:0:0 tisi0.xml
ligolw_tisi --instrument=H1=0:0:0 --instrument=H2=0:0:0 --instrument=L1=3.14159:3.14159:3.14159 --instrument=V1=7.892:7.892:7.892 tisi1.xml
ligolw_add --output $@ tisi0.xml tisi1.xml
dag : segments.xml.gz vetoes.xml.gz frame.cache tisi.xml plots $(WEBDIR) $(INJECTIONS) $(BANK_CACHE_FILES) $(FAR_INJECTIONS)
gstlal_inspiral_pipe --data-source frames --gps-start-time $(START) --gps-end-time $(STOP) --frame-cache frame.cache --frame-segments-file segments.xml.gz --vetoes vetoes.xml.gz --frame-segments-name datasegments --control-peak-time $(PEAK) --num-banks $(NUMBANKS) --fir-stride 4 --web-dir $(WEBDIR) --time-slide-file tisi.xml $(INJECTION_LIST) --bank-cache $(BANK_CACHE_STRING) --tolerance 0.9999 --overlap $(OVERLAP) --flow $(LOW_FREQUENCY_CUTOFF) $(CHANNEL_NAMES) --autocorrelation-length $(AC_LENGTH) --samples-min $(SAMPLES_MIN) --samples-max-256 $(SAMPLES_MAX_256) $(ADDITIONAL_DAG_OPTIONS) $(FAR_INJECTION_LIST)
V1_frame.cache:
# FIXME force the observatory column to actually be instrument
ligo_data_find -o V -t $(VIRGO_FRAME_TYPE) -l -s $(START) -e $(STOP) --url-type file | awk '{ print $$1" V1_"$$2" "$$3" "$$4" "$$5}' > $@
%_frame.cache:
#FIXME horrible hack to get the observatory, not guaranteed to work
$(eval OBS:=$*)
$(eval OBS:=$(subst 1,$(empty),$(OBS)))
$(eval OBS:=$(subst 2,$(empty),$(OBS)))
# FIXME force the observatory column to actually be instrument
ligo_data_find -o $(OBS) -t $(LIGO_FRAME_TYPE) -l -s $(START) -e $(STOP) --url-type file | awk '{ print $$1" $*_"$$2" "$$3" "$$4" "$$5}' > $@
frame.cache: $(FRAME_CACHE_FILES)
cat $(FRAME_CACHE_FILES) > frame.cache
segments.xml.gz: frame.cache
gsiscp pcdev3.cgca.uwm.edu:/home/channa/public_html/SELECTED_SEGS.xml.gz $@
gstlal_cache_to_segments frame.cache nogaps.xml
gstlal_segments_operations --segment-file1 $@ --segment-file2 nogaps.xml --intersection --output-file $@
-rm -vf nogaps.xml
gstlal_segments_trim --trim 8 --gps-start-time $(START) --gps-end-time $(STOP) --min-length 2048 --output $@ $@
vetoes.xml.gz:
gsiscp pcdev3.cgca.uwm.edu:/home/channa/public_html/COMBINED_CAT_4_VETO_SEGS.xml.gz $@
gstlal_segments_trim --gps-start-time $(START) --gps-end-time $(STOP) --segment-name vetoes --output $@ $@
clean:
-rm -rvf *.sub *.dag* *.cache *.sh logs *.sqlite plots *.html Images *.css *.js
-rm -rvf lalapps_run_sqlite/ ligolw_* gstlal_*
-rm -vf segments.xml.gz tisi.xml H*.xml L*.xml V*.xml ?_injections.xml ????-*_split_bank-*.xml vetoes.xml.gz
-rm -vf *marginalized*.xml.gz *-ALL_LLOID*.xml.gz
-rm -vf tisi0.xml tisi1.xml
-rm -rf *_split_bank
-rm -rf $(INJECTIONS)
gstlalcbc@pcdev3:~/MDC/NSBH/Summer2014/recolored/nonspin/966384015-967384015-tito-bank-1401$ diff -u Makefile ../966384015-967384015-tito-bank/Makefile
--- Makefile 2014-11-10 18:14:31.582722944 -0600
+++ ../966384015-967384015-tito-bank/Makefile 2014-11-07 15:40:35.044299827 -0600
@@ -10,11 +10,11 @@
IFOS = H1 L1 V1
START = 966384015
STOP = 967384015
-TAG = tito-bank-1401
+TAG = tito-bank
WEBDIR = ~/public_html/MDC/NSBH/Summer2014/recolored/nonspin/$(START)-$(STOP)-$(TAG)
NUMBANKS = 4
PEAK = 0
-AC_LENGTH = 1401
+AC_LENGTH = 351
SAMPLES_MIN = 512
SAMPLES_MAX_256 = 512
gstlalcbc@pcdev3:~/MDC/NSBH/Summer2014/recolored/nonspin/966384015-967384015-tito-bank-1401$ diff -u Makefile ../966384015-967384015-tito-bank-701/Makefile
--- Makefile 2014-11-10 18:14:31.582722944 -0600
+++ ../966384015-967384015-tito-bank-701/Makefile 2014-11-08 15:13:59.070203224 -0600
@@ -10,11 +10,11 @@
IFOS = H1 L1 V1
START = 966384015
STOP = 967384015
-TAG = tito-bank-1401
+TAG = tito-bank-701
WEBDIR = ~/public_html/MDC/NSBH/Summer2014/recolored/nonspin/$(START)-$(STOP)-$(TAG)
NUMBANKS = 4
PEAK = 0
-AC_LENGTH = 1401
+AC_LENGTH = 701
SAMPLES_MIN = 512
SAMPLES_MAX_256 = 512
The results are here:
The range plots are shown here:
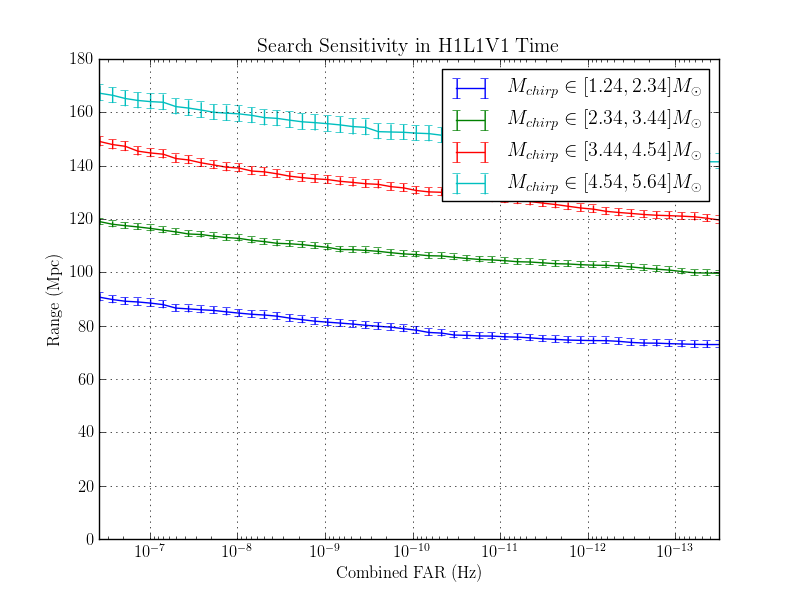
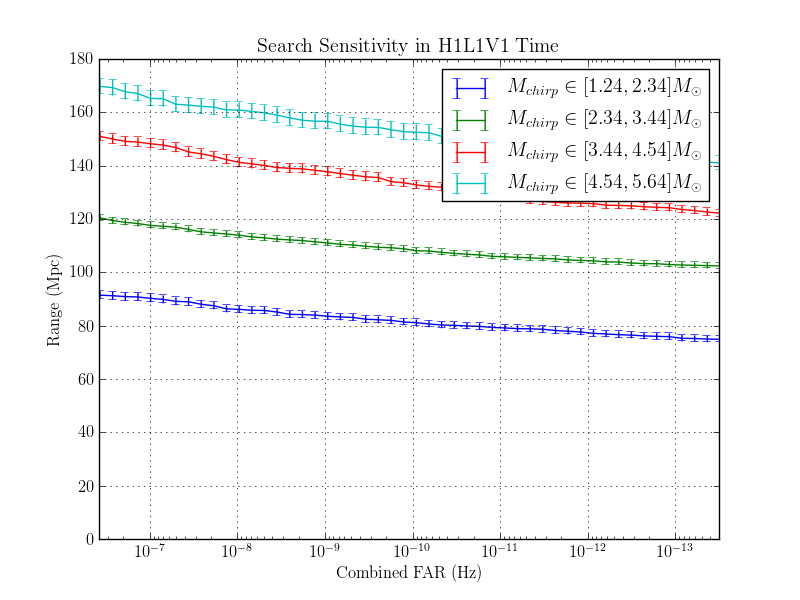
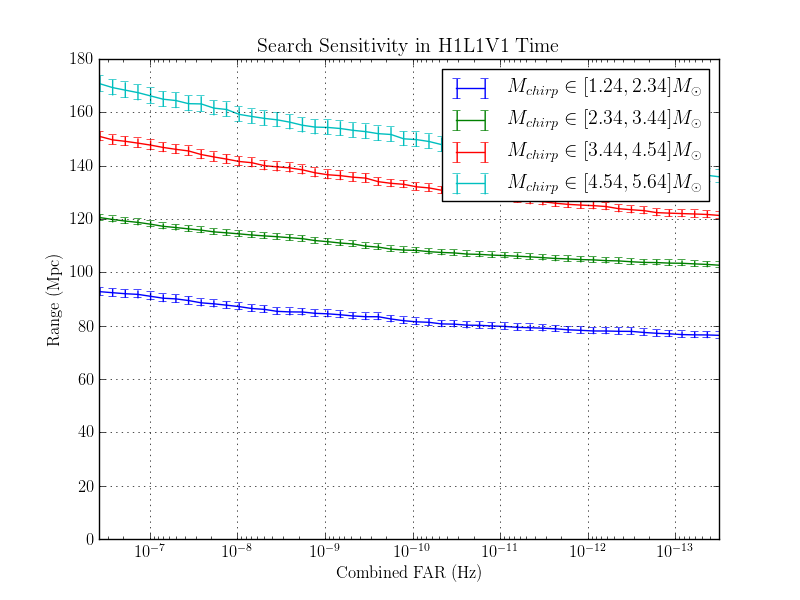
 1.8.1.2
1.8.1.2