 |
gstlal-inspiral
0.4.2
|
This page describes a study of the BNS MDC described here:
The goal is to establish if the results of this MDC are reasonable. The following aspects were checke
The following makefile was used to produce the results
#
# Template bank parameters
#
# The filtering start frequency
LOW_FREQUENCY_CUTOFF = 30.0
# The maximum frequency to filter to
HIGH_FREQUENCY_CUTOFF = 2048.0
# Controls the number of templates in each SVD sub bank
NUM_SPLIT_TEMPLATES = 100
# Controls the overlap from sub bank to sub bank - helps mitigate edge effects
# in the SVD. Redundant templates will be removed
OVERLAP = 20
# The program used to make the template bank. This will be searched for in the
# process param table in order to extract some metadata
BANK_PROGRAM = pycbc_geom_nonspinbank
# The approximant that you wish to filter with
APPROXIMANT = TaylorF2
#
# Triggering parameters
#
# The detectors to analyze
IFOS = H1 L1
# The GPS start time
START = 966384015
# The GPS end time
STOP = 971384015
# A user tag for the run
TAG = pipe-compare-CAT2
# A web directory for output
WEBDIR = ~/public_html/MDC/BNS/Summer2014/recolored/nonspin/$(START)-$(STOP)-$(TAG)
# The number of sub banks to process in parallel for each gstlal_inspiral job
NUMBANKS = 8
# The control peak time for the composite detection statistic. If set to 0 the
# statistic is disabled
PEAK = 0
# The length of autocorrelation chi-squared in sample points
AC_LENGTH = 701
# The minimum number of samples to include in a given time slice
SAMPLES_MIN = 512
# The maximum number of samples to include in the 256 Hz or above time slices
SAMPLES_MAX_256 = 512
#
# additional options, e.g.,
#
#ADDITIONAL_DAG_OPTIONS = "--blind-injections BNS-MDC1-WIDE.xml"
#
# Injections
#
# The seed is the string before the suffix _injections.xml
# Change as appropriate, whitespace is important
INJECTIONS := BNS-SpinMDC-ISOTROPIC.xml BNS-SpinMDC-ALIGNED.xml BNS-SpinMDC-ALIGNED-extrainj-EVEN.xml BNS-SpinMDC-ALIGNED-extrainj-ODD.xml BNS-SpinMDC-ISOTROPIC-extrainj-EVEN.xml BNS-SpinMDC-ISOTROPIC-extrainj-ODD.xml
# NOTE you shouldn't need to change these next two lines
comma:=,
INJECTION_REGEX = $(subst $(space),$(comma),$(INJECTIONS))
#
# Segment and frame type info
#
# The LIGO and Virgo frame types
LIGO_FRAME_TYPE='T1200307_V4_EARLY_RECOLORED_V2'
VIRGO_FRAME_TYPE='T1300121_V1_EARLY_RECOLORED_V2'
# The Channel names. FIXME sadly you have to change the CHANNEL_NAMES string if
# you want to analyze a different set of IFOS
H1_CHANNEL=LDAS-STRAIN
L1_CHANNEL=LDAS-STRAIN
V1_CHANNEL=h_16384Hz
CHANNEL_NAMES:=--channel-name=H1=$(H1_CHANNEL) --channel-name=L1=$(L1_CHANNEL) --channel-name=V1=$(V1_CHANNEL)
#
# Get some basic definitions. NOTE this comes from the share directory probably.
#
include Makefile.offline_analysis_rules
#
# Workflow
#
all : dag
BNS_NonSpin_30Hz_earlyaLIGO.xml:
gsiscp sugar-dev1.phy.syr.edu:/home/jveitch/public_html/bns/mdc/spin/tmpltbank/BNS_NonSpin_30Hz_earlyaLIGO.xml .
$(INJECTIONS):
gsiscp sugar-dev1.phy.syr.edu:/home/jveitch/public_html/bns/mdc/spin/"{$(INJECTION_REGEX)}" .
%_split_bank.cache : BNS_NonSpin_30Hz_earlyaLIGO.xml
mkdir -p $*_split_bank
gstlal_bank_splitter --f-low $(LOW_FREQUENCY_CUTOFF) --group-by-chi --output-path $*_split_bank --approximant $(APPROXIMANT) --bank-program $(BANK_PROGRAM) --output-cache $@ --overlap $(OVERLAP) --instrument $* --n $(NUM_SPLIT_TEMPLATES) --sort-by mchirp --add-f-final --max-f-final $(HIGH_FREQUENCY_CUTOFF) $<
plots :
mkdir plots
$(WEBDIR) :
mkdir -p $(WEBDIR)
tisi.xml :
ligolw_tisi --instrument=H1=0:0:0 --instrument=H2=0:0:0 --instrument=L1=0:0:0 --instrument=V1=0:0:0 tisi0.xml
ligolw_tisi --instrument=H1=0:0:0 --instrument=H2=0:0:0 --instrument=L1=3.14159:3.14159:3.14159 --instrument=V1=7.892:7.892:7.892 tisi1.xml
ligolw_add --output $@ tisi0.xml tisi1.xml
dag : segments.xml.gz vetoes.xml.gz frame.cache tisi.xml plots $(WEBDIR) $(INJECTIONS) $(BANK_CACHE_FILES)
gstlal_inspiral_pipe --data-source frames --gps-start-time $(START) --gps-end-time $(STOP) --frame-cache frame.cache --frame-segments-file segments.xml.gz --vetoes vetoes.xml.gz --frame-segments-name datasegments --control-peak-time $(PEAK) --num-banks $(NUMBANKS) --fir-stride 4 --web-dir $(WEBDIR) --time-slide-file tisi.xml $(INJECTION_LIST) --bank-cache $(BANK_CACHE_STRING) --tolerance 0.9999 --overlap $(OVERLAP) --flow $(LOW_FREQUENCY_CUTOFF) $(CHANNEL_NAMES) --autocorrelation-length $(AC_LENGTH) --samples-min $(SAMPLES_MIN) --samples-max-256 $(SAMPLES_MAX_256) $(ADDITIONAL_DAG_OPTIONS)
V1_frame.cache:
# FIXME force the observatory column to actually be instrument
ligo_data_find -o V -t $(VIRGO_FRAME_TYPE) -l -s $(START) -e $(STOP) --url-type file | awk '{ print $$1" V1_"$$2" "$$3" "$$4" "$$5}' > $@
%_frame.cache:
# FIXME horrible hack to get the observatory, not guaranteed to work
$(eval OBS:=$*)
$(eval OBS:=$(subst 1,$(empty),$(OBS)))
$(eval OBS:=$(subst 2,$(empty),$(OBS)))
# FIXME force the observatory column to actually be instrument
ligo_data_find -o $(OBS) -t $(LIGO_FRAME_TYPE) -l -s $(START) -e $(STOP) --url-type file | awk '{ print $$1" $*_"$$2" "$$3" "$$4" "$$5}' > $@
frame.cache: $(FRAME_CACHE_FILES)
cat $(FRAME_CACHE_FILES) > frame.cache
segments.xml.gz: frame.cache
# These segments come from the MDC set
gsiscp pcdev3.cgca.uwm.edu:/home/channa/public_html/SELECTED_SEGS.xml.gz $@
gstlal_cache_to_segments frame.cache nogaps.xml
gstlal_segments_operations --segment-file1 $@ --segment-file2 nogaps.xml --intersection --output-file $@
-rm -vf nogaps.xml
gstlal_segments_trim --trim 8 --gps-start-time $(START) --gps-end-time $(STOP) --min-length 2048 --output $@ $@
vetoes.xml.gz:
gsiscp pcdev3.cgca.uwm.edu:/home/gstlalcbc/public_html/H1L1-S6MDC_COMBINED_CAT_2_HWINJ_VETO_SEGS.xml.gz $@
gstlal_segments_trim --gps-start-time $(START) --gps-end-time $(STOP) --segment-name vetoes --output $@ $@
clean:
-rm -rvf *.sub *.dag* *.cache *.sh logs *.sqlite plots *.html Images *.css *.js
-rm -rvf lalapps_run_sqlite/ ligolw_* gstlal_*
-rm -vf segments.xml.gz tisi.xml H*.xml L*.xml V*.xml ?_injections.xml ????-*_split_bank-*.xml vetoes.xml.gz
-rm -vf *marginalized*.xml.gz *-ALL_LLOID*.xml.gz
-rm -vf tisi0.xml tisi1.xml
-rm -rf *_split_bank
-rm -rf BNS_NonSpin_30Hz_earlyaLIGO.xml
-rm -rf $(INJECTIONS)
The following source code was used to produce the plots below
#!/usr/bin/python
import os
import math
import sys
from pylal import imr_utils
import matplotlib
matplotlib.use('Agg')
from matplotlib import rc
rc('text', usetex=True)
import pylab
import numpy
from optparse import OptionParser
from glue.ligolw import dbtables
from glue.ligolw import ligolw
from glue.ligolw import utils
from glue.ligolw import table
from glue.ligolw import lsctables
from glue.ligolw import utils as ligolw_utils
import sqlite3
from gstlal import far
sqlite3.enable_callback_tracebacks(True)
# define a content handler
class LIGOLWContentHandler(ligolw.LIGOLWContentHandler):
pass
def get_min_far_inspiral_injections(connection, segments = None):
found_query = 'SELECT sim_inspiral.*, coinc_inspiral.combined_far, coinc_inspiral.snr, H1.snr, H1.chisq, L1.snr, L1.chisq FROM sim_inspiral JOIN coinc_event_map AS mapA ON mapA.event_id == sim_inspiral.simulation_id JOIN coinc_event_map AS mapB ON mapB.coinc_event_id == mapA.coinc_event_id JOIN coinc_inspiral ON coinc_inspiral.coinc_event_id == mapB.event_id JOIN coinc_event on coinc_event.coinc_event_id == coinc_inspiral.coinc_event_id JOIN coinc_event_map as mapC ON mapC.coinc_event_id == coinc_inspiral.coinc_event_id JOIN coinc_event_map as mapD on mapD.coinc_event_id == coinc_inspiral.coinc_event_id JOIN sngl_inspiral as H1 ON H1.event_id == mapC.event_id JOIN sngl_inspiral AS L1 ON L1.event_id == mapD.event_id WHERE mapA.table_name = "sim_inspiral" AND mapB.table_name = "coinc_event" AND injection_in_segments(sim_inspiral.geocent_end_time, sim_inspiral.geocent_end_time_ns) AND mapC.table_name = "sngl_inspiral" and H1.ifo == "H1" and L1.ifo == "L1"'
def injection_was_made(end_time, end_time_ns, segments = segments):
return imr_utils.time_within_segments(end_time, end_time_ns, segments)
# restrict the found injections to only be within certain segments
connection.create_function("injection_in_segments", 2, injection_was_made)
# get the mapping of a record returned by the database to a sim
# inspiral row. Note that this is DB dependent potentially, so always
# do this!
make_sim_inspiral = imr_utils.make_sim_inspiral_row_from_columns_in_db(connection)
found_injections = {}
for values in connection.cursor().execute(found_query):
# all but the last column is used to build a sim inspiral object
sim = make_sim_inspiral(values[:-6])
far = values[-6]
snr = values[-5]
H1snr, H1chisq, L1snr, L1chisq = values[-4:]
# update with the minimum far seen until now
this_inj = found_injections.setdefault(sim.simulation_id, (far, snr, H1snr, H1chisq, L1snr, L1chisq, sim))
if far < this_inj[0]:
found_injections[sim.simulation_id] = (far, snr, H1snr, H1chisq, L1snr, L1chisq, sim)
total_query = 'SELECT * FROM sim_inspiral WHERE injection_in_segments(geocent_end_time, geocent_end_time_ns)'
total_injections = {}
# Missed injections start as a copy of the found injections
missed_injections = {}
for values in connection.cursor().execute(total_query):
sim = make_sim_inspiral(values)
total_injections[sim.simulation_id] = sim
missed_injections[sim.simulation_id] = sim
# now actually remove the missed injections
for k in found_injections:
del missed_injections[k]
return found_injections.values(), total_injections.values(), missed_injections.values()
lsctables.use_in(LIGOLWContentHandler)
parser = OptionParser()
parser.add_option("--db", action = "append")
parser.add_option("--marg-likelihood-file")
options, filenames = parser.parse_args()
likefile = far.parse_likelihood_control_doc(utils.load_filename(options.marg_likelihood_file, contenthandler = far.ThincaCoincParamsDistributions.LIGOLWContentHandler))[0]
def get_snr(sim, like, ifo):
hdict = like.horizon_history.getdict(sim.geocent_end_time)
snr = hdict[ifo] / getattr(sim, "eff_dist_%s" % ifo[0].lower()) * 8. * (1.22 / sim.mchirp)**(-5./6)
return snr
for db in options.db:
# Get info from the analysis database
connection = sqlite3.connect(db)
xmldoc = dbtables.get_xml(connection)
db_sim_inspiral_table = table.get_table(xmldoc, dbtables.lsctables.SimInspiralTable.tableName)
injection_segments = imr_utils.get_segments(connection, xmldoc, dbtables.lsctables.CoincInspiralTable.tableName, "gstlal_inspiral", "vetoes", data_segments_name = "datasegments")
instruments_set = frozenset(("H1", "L1"))
segments_to_consider_for_these_injections = injection_segments.intersection(instruments_set) - injection_segments.union(set(injection_segments.keys()) - instruments_set)
found, total, missed = get_min_far_inspiral_injections(connection, segments = segments_to_consider_for_these_injections)
print >> sys.stderr, "%s total injections: %d; Found injections %d: Missed injections %d" % (instruments_set, len(total), len(found), len(missed))
sims = {}
sims.update([((sim.geocent_end_time, sim.mass1, sim.mass2, sim.spin1z, sim.spin2z), (far, snr, get_snr(sim, likefile, "H1"), get_snr(sim, likefile, "L1"), H1snr, H1chisq, L1snr, L1chisq, sim)) for (far, snr, H1snr, H1chisq, L1snr, L1chisq, sim) in found])
pylab.figure()
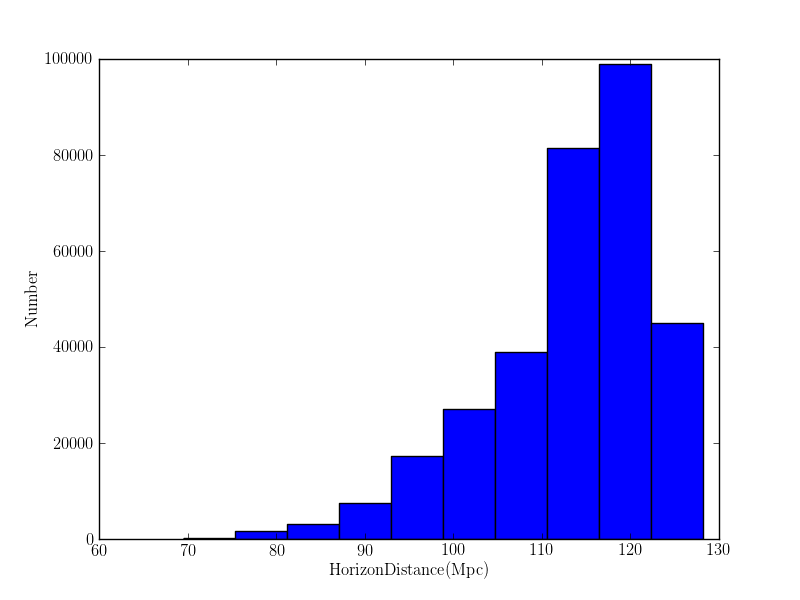
pylab.hist(likefile.horizon_history["L1"].values())
pylab.xlabel(r'$\mathrm{Horizon Distance (Mpc)}$')
pylab.ylabel(r'$\mathrm{Number}$')
pylab.savefig('/home/gstlalcbc/public_html/horizon_hist_%s' % db.replace(".sqlite", ".png"))
pylab.figure()
colors = ['b', 'g', 'r', 'k'] * 2
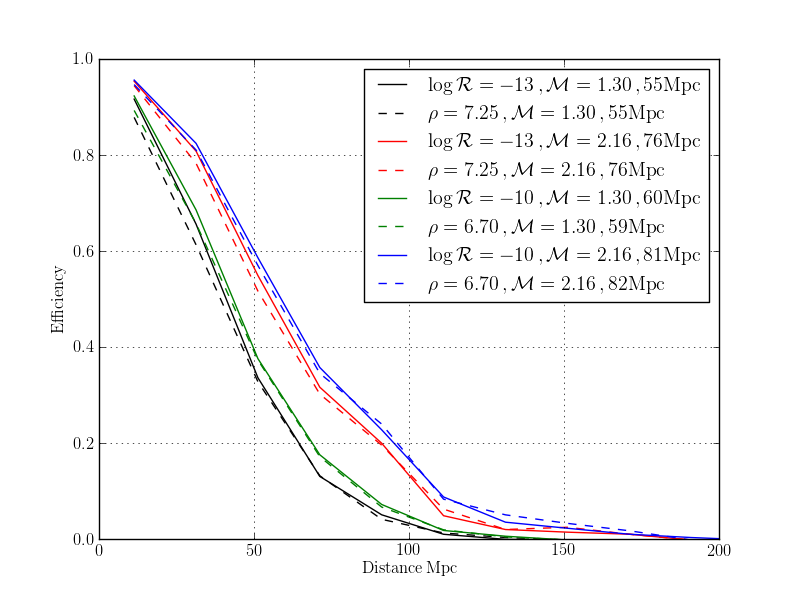
for FAR, SNR in ((1.e-13, 7.25), (1.e-10, 6.7)):
foundbyFAR = [s[8] for s in sims.values() if s[0] < FAR]
# Do a cut on expected SNR in each detector
foundbySNR = [s[8] for s in sims.values() if s[2] > SNR and s[3] > SNR]
# NOTE uncomment to plot by *Recovered* SNR
#foundbySNR = [s[8] for s in sims.values() if s[4] > SNR*.95 and s[6] > SNR*.95]
# produce two chirp mass bins and 20 distance bins
bins = imr_utils.guess_distance_chirp_mass_bins_from_sims(total, mbins = 2, distbins = 20)
effFAR, errFAR = imr_utils.compute_search_efficiency_in_bins(foundbyFAR, total, bins, sim_to_bins_function = lambda sim: (sim.distance, sim.mchirp))
effSNR, errSNR = imr_utils.compute_search_efficiency_in_bins(foundbySNR, total, bins, sim_to_bins_function = lambda sim: (sim.distance, sim.mchirp))
volFAR, verrFAR = imr_utils.compute_search_volume_in_bins(foundbyFAR, total, bins, sim_to_bins_function = lambda sim: (sim.distance, sim.mchirp))
volSNR, verrSNR = imr_utils.compute_search_volume_in_bins(foundbySNR, total, bins, sim_to_bins_function = lambda sim: (sim.distance, sim.mchirp))
for i, mchirp in enumerate(bins.centres()[1]):
color = colors.pop()
fac = (4./3 * math.pi)**(1./3)
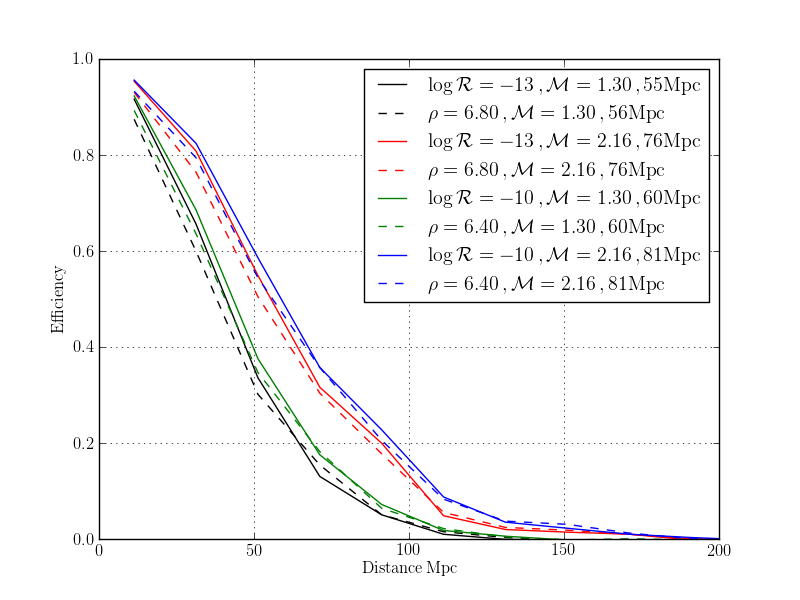
pylab.plot(effFAR.centres()[0], effFAR.array[:,i], color = color, label = r'$\log\mathcal{R}=%.0f \,, \mathcal{M} = %.2f \,, %.0f \mathrm{Mpc}$' % (math.log10(FAR), mchirp, volFAR.array[i]**(1./3) / fac))
pylab.plot(effSNR.centres()[0], effSNR.array[:,i], '--', color = color, label = r'$\rho=%.2f \,, \mathcal{M} = %.2f \,, %.0f \mathrm{Mpc}$' % (SNR, mchirp, volSNR.array[i]**(1./3) / fac))
pylab.legend()
pylab.xlim([0,200])
pylab.grid()
pylab.xlabel('$\mathrm{Distance\,Mpc}$')
pylab.ylabel('$\mathrm{Efficiency}$')
pylab.savefig('/home/gstlalcbc/public_html/efficiency_%s' % db.replace(".sqlite", ".png"))
snrx, snry, far = [],[],[]
for k in sims:
#NOTE just a maximum FAR to make the plot reasonable.
if sims[k][0] < 1e-3:
far.append(sims[k][0])
snrx.append(sims[k][1])
snry.append((sims[k][2]**2 + sims[k][3]**2)**.5)
logfars = numpy.log10(far)
logfars[logfars < -15] = -15
sortedfars = sorted([(far,x,y) for far,x,y in zip(logfars, snrx, snry)])
snrx = numpy.array([a[1] for a in sortedfars])
snry = numpy.array([a[2] for a in sortedfars])
logfars = numpy.array([a[0] for a in sortedfars])
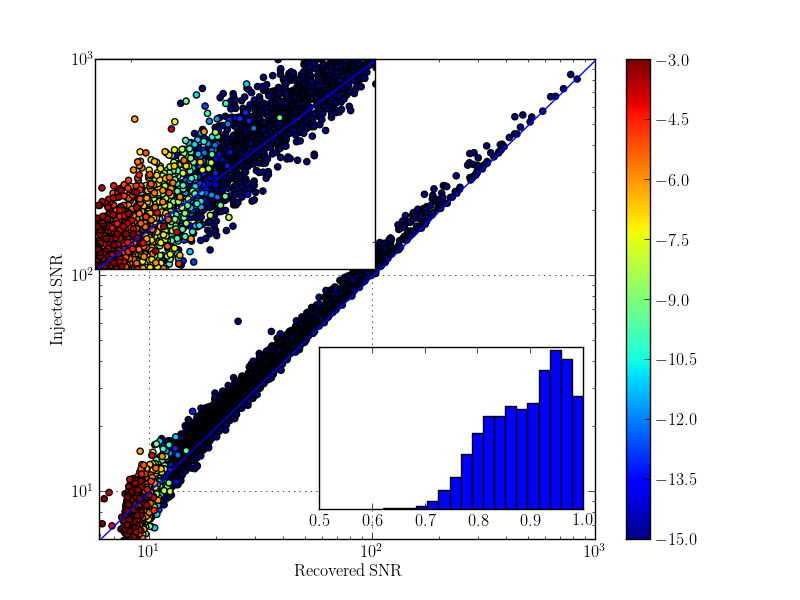
pylab.figure()
pylab.scatter(snrx, snry, c = logfars)
pylab.xlabel(r'$\mathrm{Recovered\,SNR}$')
pylab.ylabel(r'$\mathrm{Injected\,SNR}$')
pylab.loglog([5,1000], [5,1000])
pylab.xlim([6,1000])
pylab.ylim([6,1000])
pylab.grid()
pylab.colorbar()
a = pylab.axes([.12, .55, .35, .35], axisbg='w')
pylab.scatter(snrx, snry, c = logfars)
pylab.loglog([5,1000], [5,1000])
pylab.grid()
pylab.xlim([8,20])
pylab.ylim([8,20])
pylab.setp(a, xticks=[], yticks=[])
a = pylab.axes([.4, .15, .33, .27], axisbg='w')
pylab.hist([x / y for x, y, lf in zip(snrx, snry, logfars) if lf < -7], numpy.linspace(0.6,1.0,20))
pylab.setp(a, xticks = [0.5, 0.6, 0.7, 0.8, 0.9, 1.0], yticks=[])
pylab.savefig('/home/gstlalcbc/public_html/SNR_%s' % os.path.split(db)[1].replace(".sqlite", ".png"))
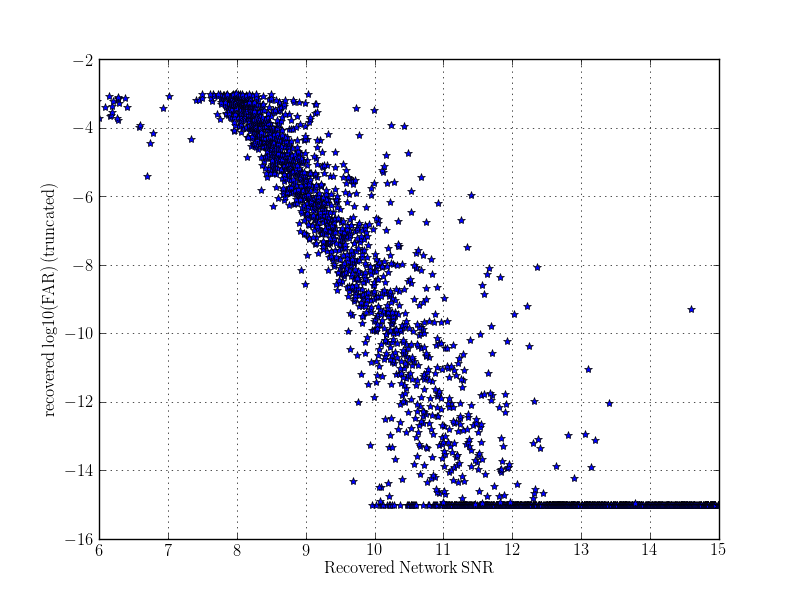
pylab.figure()
pylab.plot(snrx, logfars, '*')
pylab.xlabel(r'$\mathrm{Recovered\,Network\,SNR}$')
pylab.ylabel(r'$\mathrm{recovered\,log10(FAR)\,(truncated)}$')
pylab.xlim([6, 15])
pylab.grid()
pylab.savefig('/home/gstlalcbc/public_html/FAR_%s' % os.path.split(db)[1].replace(".sqlite", ".png"))
And it is invoked like this:
./gstlal_inspiral_injection_snr --db H1L1-ALL_LLOID_BNS_SpinMDC_ALIGNED-966384015-5000000.sqlite --marg-likelihood-file marginalized_likelihood.xml.gz
A sensible question to ask is, what false alarm rate do we expect from Gaussian noise with a single detector SNR threshold of 6.8 in each of two detectors. The following python code attempts to predict this:
#!/usr/bin/python from scipy.stats import chi2 # NOTE assume that the autocorrelation width is about 100 samples auto_correlation_time = 100. / 4096 independent_rate = 1. / auto_correlation_time live_time = 2.5e6 SNR = 6.8 window = 0.01 # Number of independent templates of the 8000 input templates # NOTE we see roughly a factor of 10 more true templates than independent # templates when trying to measure this template_trials = 8000 * 0.1 prob = 1. - chi2.cdf(SNR**2, 2) rate = prob * independent_rate joint_rate = rate * rate * window joint_prob = joint_rate * live_time joint_prob_after_trials = template_trials * joint_prob
The answer is 3e-10 which we should compare with the 5 sigma threshold of the pipeline, i.e., 5e-7. Thus the pipeline reports a false alarm probability that is approximately 1000 times higher than expected in Gaussian noise at an effective sngl detector threshold of 6.8
 1.8.1.2
1.8.1.2